
Combining in vivo genetic perturbation with CRISPR screening
An interview with Dr Randall Platt, Professur Biologisches Engineering, ETH Zurich, conducted by April Cashin-Garbutt, MA (Cantab)
Recent progress in genome editing has provided a suite of technologies enabling the rapid and flexible testing of molecular hypotheses for connecting genotype to phenotype in meaningful model systems. In the following interview, Dr Randall Platt outlines the impact of CRISPR-Cas9 and also discusses the future of the technology through combining in vivo genetic perturbation with genetic screening.
What is the CRISPR-Cas9 system?
CRISPR, which stands for Clustered Regularly Interspaced Short Palindromic Repeats, is an interesting phenomenon that occurs in prokaryotic organisms (such as bacteria) and is the basis for adaptive immunity. Adaptive immunity is the ability for your body, or a cell, to learn about foreign invaders such as viruses, and defend against these threats.
CRISPR, and associated nucleases (CRISPR-Cas), provide immunological memory and defence against invading elements. The CRISPR-Cas system is modular in the sense that it has a suite of proteins that enable storage and immunological memory (CRSIPR adaptation), creation of CRISPR guide RNAs that are complementary to threats (biogenesis), and finally detection and interference against threats (interference).
The last step, interference, is where Cas9 comes in. Cas9 is the nuclease that uses the CRISPR guide RNA to search for, detect, and destroy threats. Thus Cas9 is a kind of adaptive immune interfering protein.
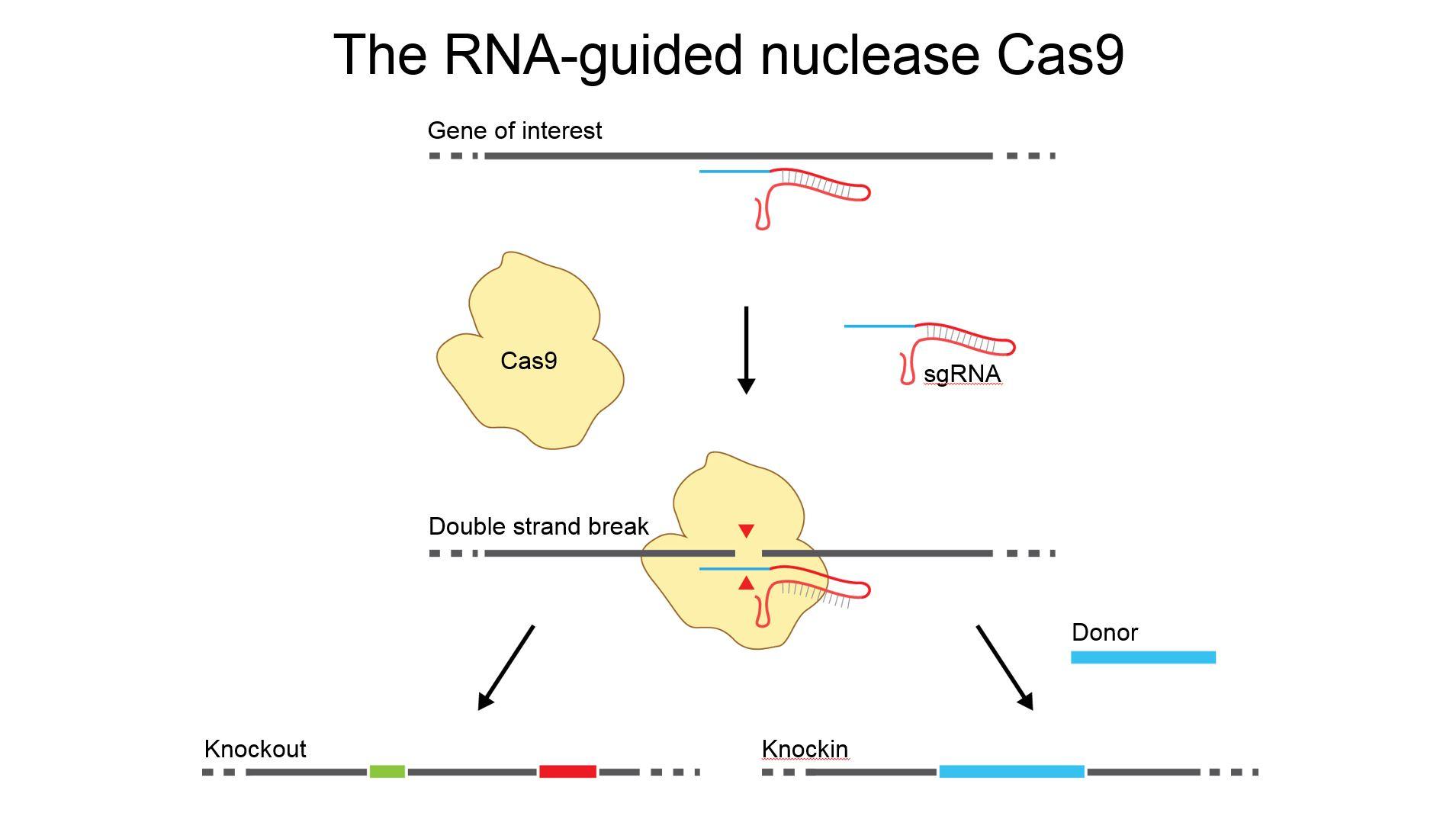
Why has CRISPR-Cas9 had such a significant impact on biology?
A few critical people in the early understanding of this technology realized that the CRISPR-Cas9 system could be leveraged as a gene editing tool. They found that this immunological memory portion of the CRISPR RNA could be reprogrammed to target any site.
This discovery was very impactful because although there were already technologies that existed to perform gene editing, they were cumbersome, or expensive, and had a number of limitations. This new technique was very interesting because for each new target gene, you just had to synthesize a piece of RNA and the rules for designing it were very trivial.
CRISPR brought genome editing to the people, as it made it possible for everybody to engineer their cells. It didn’t take that much extra expertise, beyond molecular and cellular biology, so CRISPR democratized the use of genome editing throughout the research community.
Furthermore, given CRISPR’s efficiency, it also revived the gene therapy field and it’s raised quite a bit of excitement for correcting genetic disorders where classical gene therapy, or other gene editing technologies, were not sufficient.
But I would say the major impact is that CRISPR has made genome editing possible for anyone and everyone.
How can CRISPR-Cas9 be applied to the study of genetic disorders?
CRISPR-Cas9 can be applied in a few ways, one of which is for modelling genetic disorders. If a disorder is genetic, and you know what the genes are, CRISPR can be used to recreate those genetic mutations in cells or animals in order to better understand the underlying mechanism of the disorder and also test therapeutics.
Furthermore, CRISPR-Cas9 can be applied directly to correct mutations that cause disease. The concept here is straightforward, whereby if a single mutation causes a disorder, correction of that mutation should be therapeutic if corrected in the relevant cells at the appropriate time. Thus CRISPR can be used as a therapeutic tool.
There are also ways to understand genes that modify genetic disorders. The genetic disorders that we know most about generally have only one mutation, and CRISPR-Cas9 can be used to identify for “genetic modifiers”, which are genes that, when mutated, make the prognosis better or worse. This can help you better understand the mechanism of the disease, and also come up with potential drug targets.

Please can you outline your recent work combining in vivo genetic perturbation with CRISPR screening?
A few years ago, my colleagues and I engineered a mouse that possesses this CRISPR-Cas9 protein, which made mutating genes in a living animal incredibly easy. A key benefit of being able to mutate genes directly in an animal, in contrast to mutating cells grown in culture on plastic, is that the cells in a living animal better represent their true biological state. One application of these mice that I am particularly excited about is leveraging them for genetic screens.
A genetic screen is an approach that allows one to identify genes that underlie a phenotype (e.g., cell proliferation, cell death, fate specification, etc), which is achieved by mutating many genes and then determining which gene when mutated leads to the phenotype of interest. This is conducted such that individual cells only get a single mutation. For the most part, genetic screens have only been performed in cells grown in culture (i.e. a test tube), and therefore this approach allows us to identify genes underlying phenotypes and a highly relevant context.
So far, we’ve successfully done this in proof of concept experiments to reveal important tumour suppressors for glioblastoma and hepatocellular carcinoma and, more recently, we have been applying it to interrogate master regulators of cortical development.
How have you used CRISPR to study cortical development?
We have been applying the in vivo genetic screening methodology developed for identifying tumour suppressor genes to identify master regulators that orchestrate the development of the cortex. Our approach is to introduce CRISPR-Cas9 reagents early on in embryonic development such that genes that are potentially important for cortical development are mutated in cortical progenitor cells. This is done in such a way that we mutate on the order of hundreds to thousands of genes in a single developing embryo, where any given cell only has a single mutation. Then, after the brain has developed we sort specific cell types and determine which gene was important for its development.
Do you think it will be possible one day to use CRISPR to elucidate causal variants at a pace that can match other methods of gene discovery?
Yes and no. I think it can match the pace when there are clear and meaningful cellular phenotypes. For example, in the context of cancer, the cellular phenotype of proliferation is both clear and meaningful for the disease pathology and can be easily investigated in single cells. But, on the other hand, if you’re talking about neuro-development, or neuro-psychiatric disorders, where the phenotypes are much more subtle, or based on behaviour, this type of work generally still relies on the hard work of neuroscientists and neuro-biologists.
How important is distinguishing causal variants for advancing personalized medicine?
The way I think about it is that it is less important to find the causal variants, than meaningful variants that inform about particular patient cohorts. It doesn’t matter if a variant is causal or not, if you can say that this mutation defines a particular class of patients and can be leveraged for diagnostic or therapeutic purposes.
What are the main hurdles that still need to be overcome?
In terms of CRISPR itself, the main limitation is that we cannot precisely change DNA. We mostly rely on random DNA repair outcomes, which is fine for knocking out a gene, but if we want to make specific mutation, it is a huge hurdle.

About Dr Randall Platt
Randall Platt studied biomedical engineering and chemistry at the University of Utah in Salt Lake City, Utah. In 2011 he obtained an MPhil from Imperial College London in material science and in 2015 a PhD from MIT in biological engineering.
After a postdoctoral fellowship at the Broad Institute of MIT and Harvard jointly between Feng Zhang and Paola Arlotta, Platt was appointed as a tenure-track assistant professor in the department of Biosystems Science and Engineering (D-BSSE) at ETH Zurich in 2016.
The Platt group develops and applies molecular tools for applications at the intersection of genome engineering, synthetic biology, and neurobiology.