Unlocking neural diversity: A new tool for multimodal profiling of brain cell types
The mammalian brain contains billions of cells, of thousands of different types. Each type, with unique properties and functions, can be defined by its gene activity, or expression. But understanding the brain requires more than just cataloguing its components. It needs a way to link multiple layers of information – from a cell’s structure to its connections to its electrical firing patterns.
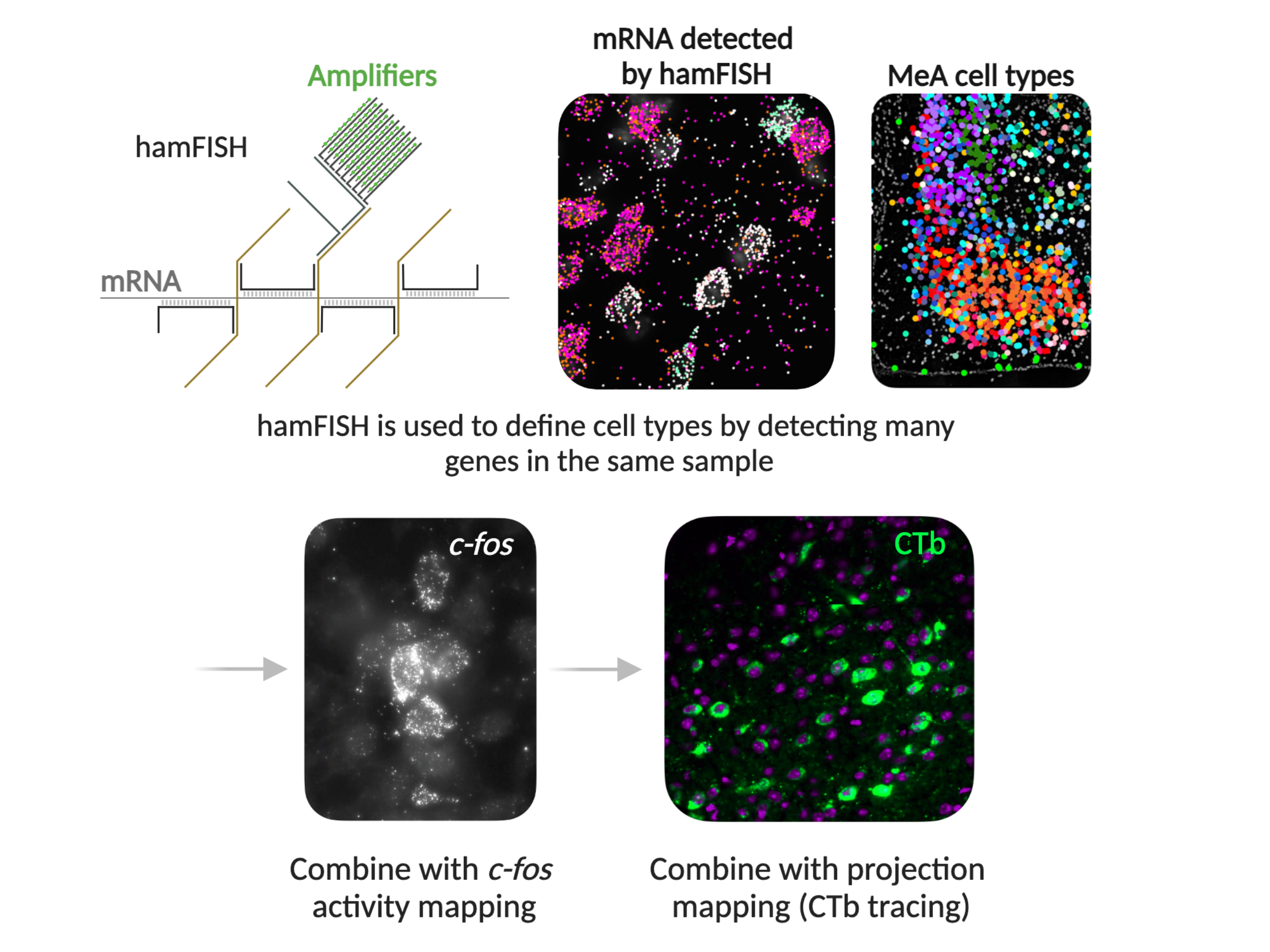
These layers of complexity inspired SWC scientists to develop hamFISH (highly amplified multiplexed fluorescence in situ hybridisation). This novel tool allows researchers to identify which genes are expressed within individual cells while preserving the tissue architecture. Designed to streamline multimodal profiling of neural cells, hamFISH can be combined with techniques to simultaneously study cell connectivity with other brain areas and activation after specific animal behaviours.
Dr Mathew Edwards, Research Fellow at the SWC, led the development of hamFISH together with SWC Group Leader Dr Yoh Isogai, now based at the Allen Institute for Neural Dynamics in Seattle.
The team used hamFISH to uncover the locations of different cell types in the mouse medial amygdala (MeA), an area that controls innate social behaviour, for the first time. Their findings have recently been published in eLife.
The need for multimodal profiling
Neuroscientists are increasingly looking to connect the dots between different aspects of neurons — their connectivity, structure, functions, and gene expression.
A choice of methods can determine gene expression within a cell, and therefore its type. Single-cell RNA sequencing methods reveal the expressed genes, but they destroy spatial information in the process. In contrast, imaging-based techniques to determine gene expression preserve tissue architecture but are often laborious, require specialist complex analysis and are expensive.
“We wanted to make this process easier to do in a regular laboratory. We wanted anyone to be able to define cell types in their area of interest,” says Mathew Edwards, Research Fellow at the SWC.

What is hamFISH?
hamFISH enables the sequential detection of expressed genes in brain tissue through multiplexed branched DNA amplification. After tissue preparation and staining, coverslip-mounted brain sections are placed in a flow chamber on a microscope, where reagents containing fluorescently-labelled readout probes can pass directly across the sample. The team imaged the signal from 3 genes at a time using green, red and far-red dyes, across 11 rounds of staining using a fully-automated imaging pipeline.
The researchers tested the technique on the MeA, a brain region crucial for innate social behaviours. They chose 31 genes to study, based on published data from the Allen Mouse Brain Atlas and others.
As with similar technologies, the challenge in developing hamFISH was overcoming background fluorescence from brain tissue and non-specific signals, to detect the specific signals of interest from the chosen genes. The team achieved this using several steps. First, they included a tissue clearing stage, to remove lipids, proteins and other debris from the cells. They also included a bridge probe, meaning that two probe molecules, instead of one, have to stick to their target to act as a binding site for amplification. Both of these developments increase the fluorescent signal that can be detected in the readout and decrease non-specific fluorescence.
Implications of hamFISH
Mathew says that thanks to the high amplification, the technique should work on other regions of the brain, other organs, or even in other species.
“The cool thing is we’ve developed a ‘version 2’ which gives you an even bigger amplification. We are really hoping this technology will allow people to do these experiments in human tissue – which has a high level of background noise,” he said.
“One big advantage of hamFISH is that you can do it pretty quickly compared with previous methods. This means you can increase throughput to more readily combine it with connectivity and cell activity readouts to get statistically significant numbers” says Mathew.
Profiling the medial amygdala
Altogether, the team profiled 643,834 cells from the MeA from 49 samples.
The MeA region is crucial for innate social behaviours such as aggression, mating, and parenting in mice. The team identified 16 inhibitory and 10 excitatory neuronal types in the MeA, many of which were spatially clustered.
“The first thing we noticed was the really striking spatial patterns of the cell types. Before this, we only really knew where a few cell types were in the amygdala,” said Mathew.
The team also discovered a rare, inhibitory cell type, that was only present in the outer layer of the MeA.
They then combined hamFISH with MeA output mapping to understand if certain MeA cell types project their axons to other brain regions involved in social behaviour. They found cell-type biases in projections to the medial preoptic area (MPOA), bed nucleus of the stria terminalis (BNST), and the ventrolateral portion of the ventral medial hypothalamus (VMHvl).
Finally, they combined hamFISH with c-fos activity mapping to study the activation of the neurons in different social situations including when males interacted with another male, a female or pups.
They found that most of the cell types were activated in combinations, rather than one cell type being activated in one social situation. There was overlap between the cell types active when animals were parenting and partnering, for example. This generates unique MeA cell population activities.
“This was surprising,” says Mathew. “And interesting, because it means that actually many cell types are active during different behaviours. It’s now a question of trying to understand how the MeA uses this information at the circuit level to control social behaviour.”

A versatile tool for neuroscience
The hamFISH profiling of the MeA provides a proof-of-principle that gene expression data can be overlayed with connectivity information and activity in a behaviourally relevant way.
“My advice for anyone wanting to use this new tool is to pick your genes of interest carefully. Use as many resources as possible to make sure you are including genes that will be most useful in determining cell types.”
As neuroscience continues to push toward integrating multimodal data, tools like hamFISH will play a critical role in advancing our understanding of the brain’s complexity.