
Peeking inside the black box with recurrent neural networks
An interview with Dr. Kanaka Rajan, Harvard University, conducted by Hyewon Kim
How do we extract essential features from the activity of billions of neurons in the brain? In a SWC Seminar, Dr. Kanaka Rajan shared her work using recurrent neural networks constrained directly by experimental recordings, a special class of artificial neural network models, to figure out how real brains learn. In this Q&A, she expands on how her models reflect excitatory, inhibitory, and recurrent connections in the brain that cannot be obtained only through measurements, her most exciting finding so far, and more.
What first drew you to study neural dynamics and theories that bridge neurobiology and artificial intelligence?
My route to computational neuroscience was not a straight line. As a young student in India, I followed the typical academic path: studying math, physics, and engineering. My interest in neuroscience arose after my undergraduate degree when I took an internship position related to mental health research. I was excited by this novel environment, but it also felt familiar because my grandmother—who helped raise me—lived a challenging life with a schizophrenia diagnosis. Following this experience, I decided to study neuroscience in graduate school in order to make a difference in brain health. After my doctoral and postdoctoral training, equipped with tools from the physical sciences and my early education in computational approaches, I was uniquely positioned to choose within the realm of biology and investigate it with a computational lens. This is what my research program focuses on today: learning how we learn by bridging neurobiology with artificial intelligence.
Why do we need to extract essential features from large-scale neural data?
The brain is an incredibly complex organ with billions of neurons, and large-scale neural data allows us to capture this complexity. To quickly and accurately analyze this data in search of valuable patterns, we must first cut through the overwhelming amount of information. Extracting essential features helps us identify shared principles and mechanisms underlying brain function and enables generalizations across different experimental conditions, species, or brain regions.
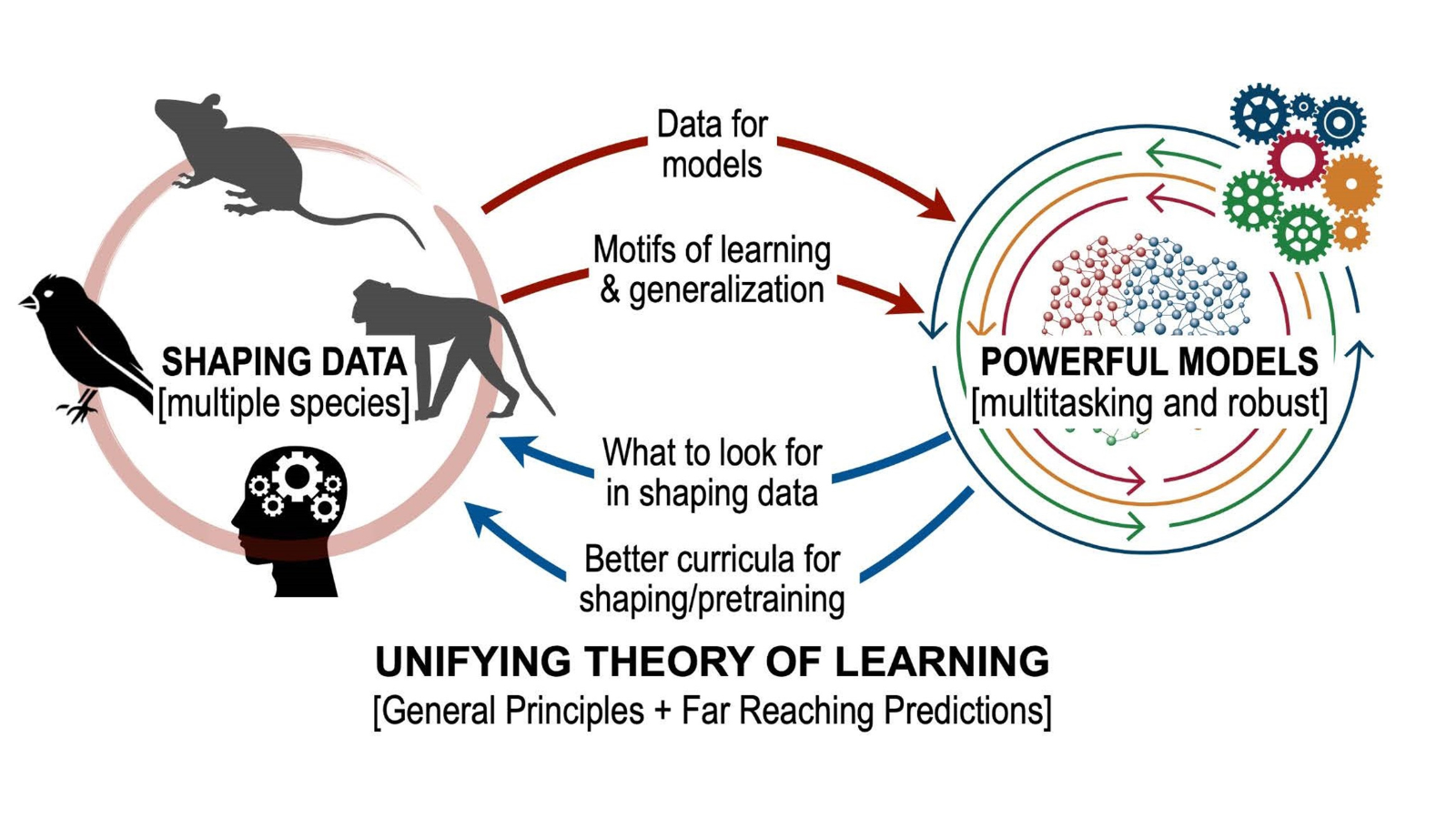
You described inputs as the ‘why’ and the ‘how’, rather than the ‘what’ of outputs. How much has been known historically about excitatory and inhibitory input currents? What about how multiple brain regions interact? How do your models reflect this?
Most animals have anatomical modularity within the brain (brain regions, cortical layer, circuits, etc.), and most modern neuroscience research seek to understand fundamental principles of the brain by using a similarly compartmentalized perspective (i.e., What brain circuits are responsible for X, Y, or Z?). However, such a simplified view of the brain does account for the myriad and complex ways in which different regions are connected via excitatory or inhibitory connections, with a large number of recurrent connections between them. Because of this reciprocal and intricate architecture, brain regions should not be considered independent nodes as they have been investigated in the past. Instead, they form an extremely complex and heterogeneous ‘network of networks’ to translate information from the world into cognition and behavior.
My work attempts to uncover how multiple brain regions interact and to dissect the brain-wide interplay of excitatory and inhibitory currents. Units in my models are analogous to neurons, integrating information from many sources through weighted connections and output rules governed by nonlinearities. By studying a population of these units, we can extract information and make decisions about the nature of the inputs driving the network. Recurrent neural networks (RNNs) are a special class of artificial neural network models where each unit both outputs to and receives input from all other units in the network. RNN models of a single brain region have emerged as a powerful tool for studying neural computation, elucidating potential ways neural circuits can generate the desired dynamics. Further, since the precise connectivity and dynamics of the model are known, researchers can peek inside the ‘black box’ that transforms the known inputs into the behavioral output to reverse-engineer the properties of the network. This approach allows us to generate or test potential hypotheses for future experiments.
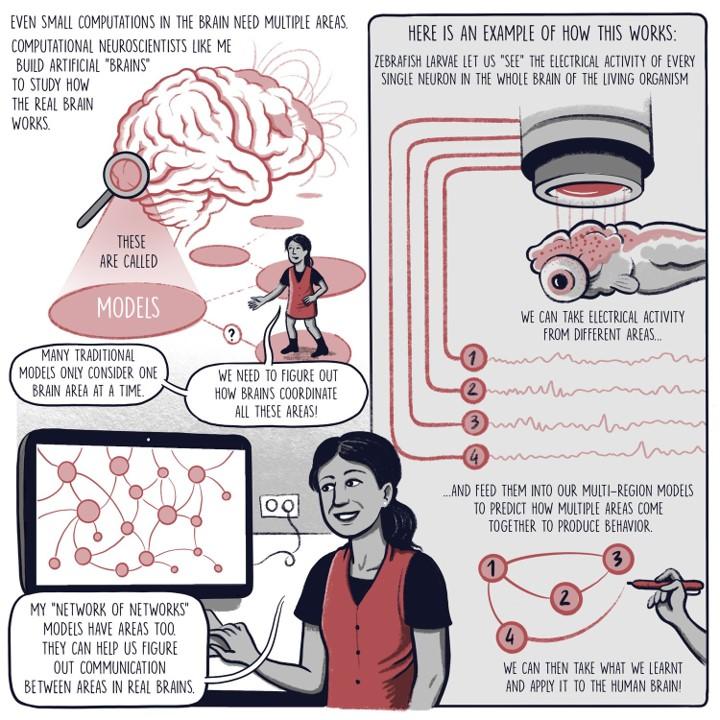
Credit: Comic designed by Matteo Farinella
Could you please describe Current-Based Decomposition (CURBD) of population activity and the role it plays in your modeling?
CURBD is a method for dissecting the complex dynamics of neural populations and uncovering the underlying mechanisms of brain function. It involves breaking down the collective activity of these neurons into different parts based on the underlying electrical currents that cause the activity. We measure the signals from many neurons simultaneously and then separate the activity into various components, each representing a specific type of current. These currents tell us about things like how neurons communicate with each other or how they generate patterns of activity. We use our CURBD to model the brain as a recurrent ‘network of networks,’ validating simulations where ground truth inter-region currents are known to untangle multi-region interactions from a range of neural datasets. CURBD helps us infer functional interactions across brain regions, from calcium signaling in mice to human memory retrieval.
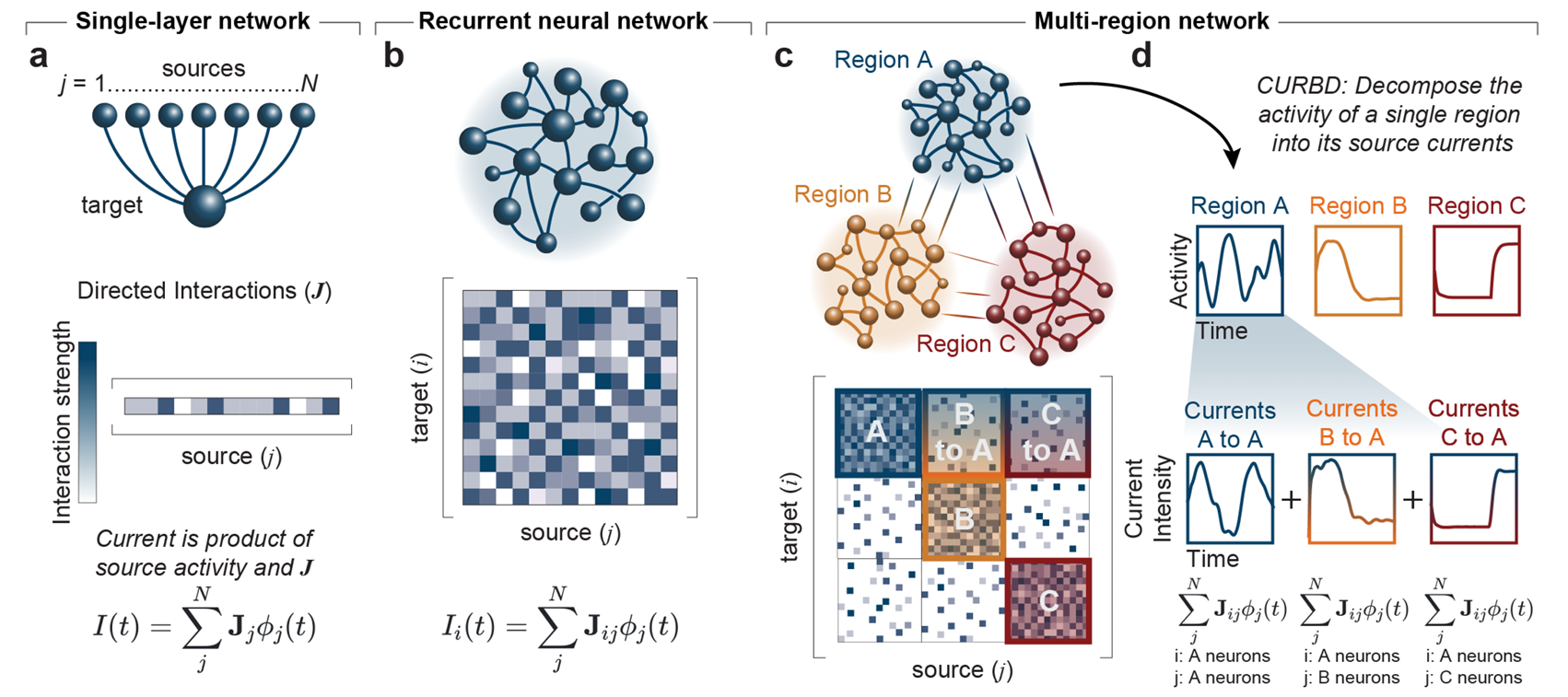
What is a key finding of yours that you find most exciting?
We recently used curriculum learning in RNNs to figure out how real brains learn! An interesting thing that happens with curriculum training is that artificial networks can begin to intuit the correct answer to a problem without having to make detailed or energy-intensive calculations. The system can start to have a “feeling” about which one is correct, almost instinctive learning at the deeper levels of the neural network. This is where our approach helps us gain novel insights: If you’re only measuring behavioral output, there are many instances in which two models can result in the same behavior, but it’s hard to know if they use the same process to get there. Discerning the correct order of operations is one of the unknown parts of the work, but seeing different models trained on different curricula come to the same conclusion offers more possibilities for the potential of human mental processes. We have shown that we can accurately predict which type of learning rule was used by an RNN based on the speed of curriculum completion. As our methods only require behavioral data as inputs, these findings open many doors for researchers in neurobiology, psychology, and beyond.
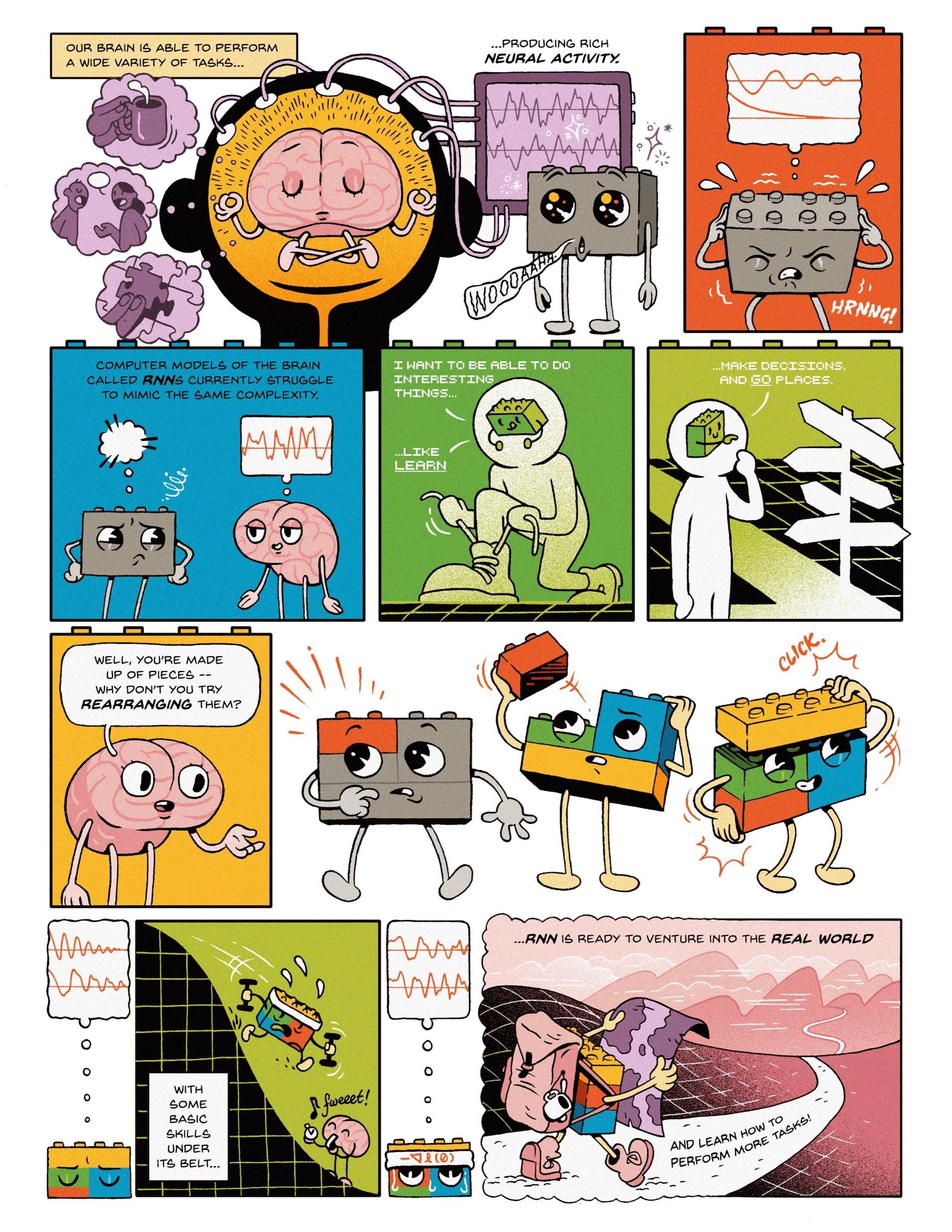
Credit: Comic designed by Jordan Collver
What is an unexpected finding that was possible with the help of your model? Can you share a bit about compositional modes and why you use them?
Organisms dynamically and continually evaluate the internal states of their brain-wide neural activity with incoming sensory and environmental evidence to decide when and how to behave. We exploit many evolutionary innovations, chiefly modularity—observable at the level of anatomically-defined brain regions, cortical layers, and cell types, among others—that can be repurposed in a compositional manner to enable us with highly flexible behavioral repertoires. Accordingly, behavior has its own observable modularity, but rarely does this behavioral modularity directly map onto traditional notions of modularity in the brain.
Compositional modes is a framework to identify the overarching compositional structure spanning specialized submodules (i.e., brain regions), allowing us to directly link the behavioral repertoire with distributed patterns of population activity brain-wide. We have applied this to whole-brain recordings of larval zebrafish with our collaborators in Dr. Karl Deisseroth’s lab at Stanford to reveal highly conserved compositional modes across individuals despite the spontaneous nature of the behavior. Thus, our results so far demonstrate somewhat unexpectedly that behavior in different individuals can be understood using a relatively small number of neurobehavioral modules—the compositional modes—and elucidate a compositional neural basis of behavior.
How does your approach differ from other approaches in studying the neural underpinnings of behavior?
While animals in neuroscience laboratories are commonly trained using shaping, few labs examine and publish data on the effectiveness and duration of training under different curricula. Our findings emphasize the importance of systematically collecting and organizing data during shaping. We demonstrate that curriculum learning in RNNs can serve as a valuable tool for investigating and distinguishing the learning principles employed by biological systems. This approach can provide insights far beyond traditional statistical analyses of learned states.
Can your key findings be applied to clinical settings? What about to other model organisms or humans?
Our models can predict where to record from, what to expect, and when abstract knowledge might emerge. If there’s an experiment where behavioral or neural data has been collected during shaping on complex time-varying tasks, it’s a pretty safe bet that our models could be applied. The more opportunities we have to test and iterate on these models, the more we will understand where (or if!) learning rules are bounded by model organisms or research settings.
What is the next piece of the puzzle you plan to focus on?
I view my work as building tools for a deeper inquiry into how humans and animals extract information from our world - with very few examples - and then apply it to new contexts. To that end, I’m excited about creating the best, most-accurate toolkits and working with collaborators to test how widely these approaches can be applied. We’ll be narrowing in on aligning the methods theorists use to construct networks with the processes responsible for task learning in biological neural circuits.

About Kanaka Rajan
Kanaka Rajan, Ph.D. is an Associate Professor in the Department of Neurobiology, Blavatnik Institute, Harvard Medical School and Faculty at the Kempner Institute for the Study of Natural and Artificial Intelligence at Harvard. Her research seeks to understand how important cognitive functions — such as learning, remembering, and deciding — emerge from the cooperative activity of multi-scale neural processes. Using data from neuroscience experiments, Kanaka applies computational frameworks derived from machine learning and statistical physics to uncover integrative theories about the brain that bridge neurobiology and artificial intelligence. For more information about Dr. Rajan, please visit www.rajanlab.com.


