
The science of wakefulness
An interview with Professor Hiroki Ueda, University Tokyo / RIKEN, conducted by Alison Cranage.
Most animals on Earth, even those without a brain, sleep. Yet the functions of slumber remain controversial and its molecular mechanisms have been elusive. Professor Hiroki Ueda, based at the University of Tokyo and RIKEN, has been at the forefront of sleep research for over 20 years. In 2016, he found that calcium signalling, known to control diverse functions in the body, also regulates sleep. In his recent SWC seminar, he discussed his work, as well as some of the methods his team has developed to enable their breakthroughs.
What brought you to study circadian rhythms and the biology of sleep?
When I started my research career I was interested in both medicine and physics. I wanted to start with the very basic things like time and space. When I looked at the literature in life sciences, time is not well defined – which I found captivating. Ageing is very interesting; a transition between states.
On a shorter timescale, you can see phenomena that oscillate, like the circadian clock that repeats roughly every 24 hours. Most biochemical reactions are sensitive to temperature, but there is a famous and interesting phenomena called temperature compensation, which allows animals to maintain this robust 24-hour rhythm independent of temperature. This was the puzzle that caught my attention.
In 2009 we identified the process that determines the 24 hours in the mammalian circadian clock, independent of temperature. It is a beautiful biochemical system.
More recently, you have uncovered a mechanism behind sleep regulation. Could you tell us about it, and the role of calcium in sleep?
After the circadian clock finding, I started to think about the next steps, including sleep regulation. This was back in 2010, and at the time, no one had a clue how sleep is regulated.
Generally, it was thought there was something missing – a ‘sleep substance’. But there were many experiments, our own and others, that couldn’t find it. Removing the related molecules didn’t give rise to a marked effect, for example.
It was in discussion with my postdoc at a conference in 2012 that I came up with the idea that there might not be a sleep substance missing, but a wakefulness one.
To test this, we created a computational model to reproduce ‘slow-wave’ patterns of neuronal activity – the state that neurons are in when we sleep. The existing models were huge and difficult to work with, so we took a theoretical physics approach and reduced it down to a model of a single neuron.
The model predicted signalling was important. Building on Dr. Setsuro Ebashi's classical discovery of calcium as a signalling molecule, we explored the hypothesis that calcium, while stimulating neurons, might also contribute to sleep promotion.
Testing our theory experimentally, we identified seven genes that work in the same calcium-related pathway to control sleep duration in mice. All seven genes allow calcium-dependent changes in neurons that make them resist becoming active.
We went on to identify the role of calcium-dependent enzymes, particularly CaMKIIα and CaMKIIβ, that promote sleep. Analysis revealed three groups of phosphorylation sites in CaMKII that are critical for sleep induction, sleep maintenance, and sleep cancellation in mice.
Further genetic screening has revealed enzymes involved in regulating cellular activities, specifically a sleep-inhibiting kinase, and sleep-promoting phosphatases, which modulate both sleep quality and quantity.
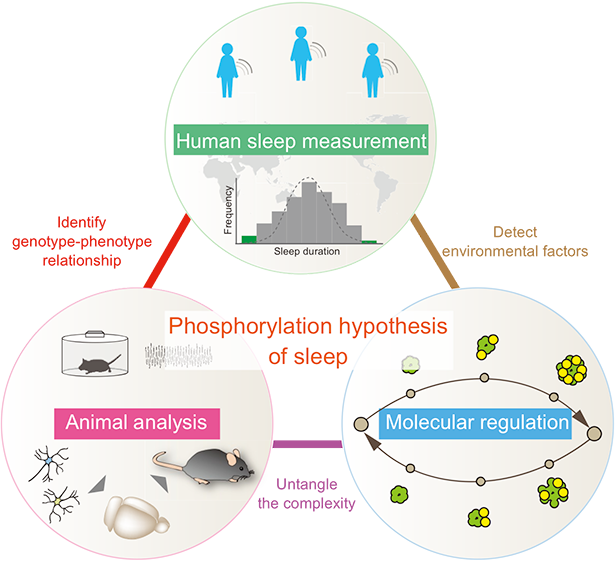
Can you expand on how you tested the computational model’s predictions in the real world?
Testing our calcium hypothesis required the development of "next-generation genetics," which is creating animals with specific genetic changes, but does not involve traditional breeding. We developed the Triple-CRISPR method in 2016, which enabled us to generate the desired mice, to study the relevant genes, within a single generation. Using this, we created 25 knockout mouse lines targeting calcium channels and pumps. Our findings with these animals showed that calcium plays a role in promoting sleep.
What we really wanted – it was the dream – was a technique to look at every single cell in the brain. And so we developed a new tissue-clearing and whole-brain cell profiling technique, called CUBIC. It was the work of three scientists in collaboration, one with a background in medicine, one in chemistry, and one in informatics. CUBIC allowed us to demonstrate that calcium can suppress neuronal excitability.
What are the next steps for your research?
Sleep is a relatively active process, even compared to ‘quiet wakefulness’, for example. Your brain cells synchronise with each other during sleep, which seems to be important for strengthening synapses, meaning your memory will be enhanced. Sleep is such an important process. In Japan, we have the world’s worst sleep habits. It’s a big issue.
We’re currently collecting sleep data in Japan. We recently developed a new technique to measure the sleep-wake cycle by using the accelerator in a smartwatch. We have 1,000 devices being used by school pupils across Japan, to study children’s sleep patterns. We can already see it’s not good enough and well below the recommendation given by the Japanese Sleep Society.
We are also studying data from UK Biobank – a repository of a huge amount of health and lifestyle information. As humans can describe themselves, how they perceive their sleep, and how they feel, there are a lot of interesting opportunities. We are studying individual differences both in terms of the genome and developmental environments.
There are many interesting correlations between the genome and human sleep behaviour. We already have a couple of gene candidates that might be involved in human sleep.
You mention synapses, which leads us to WISE (Wake Inhibition Sleep Enhancement), your newly described mechanism for the role of sleep. What does WISE propose?
The WISE mechanism suggests that periods of quiet wakefulness suppress synaptic connections, while deep sleep serves to strengthen them.
According to the model, molecules associated with synaptic suppression during prolonged quiet wakefulness, promote deep sleep. I see it as a complement rather than a contradiction to the existing theory - the synaptic homeostasis hypothesis.
The WISE mechanism also provides new insights into why chronic sleep deprivation can lead to depression, and it helps explain why rapid-acting antidepressants universally increase delta power during sleep.
This hypothesis may also clarify phenomena that existing theories struggle to address, such as the increased delta power observed after hibernation, which is associated with decreased synaptic strength.
Our results suggest that synaptic dynamics depend on maximum firing rates, not average firing rates. This provides a unified framework for the synaptic dynamics of neural networks during the sleep-wake cycle.
How close are we to getting a full picture of sleep regulation?
Hopefully, within a couple of years, we’ll have filled in the missing molecules in the calcium-dependent pathways of sleep. There are a few regulators to uncover and I’ll be working on those.
Beyond that, there are many questions about how and why neurons synchronise and desynchronise with each other. We still don’t know the basic mechanisms for that. The molecular or cellular mechanism for wakefulness and maybe consciousness might be a challenge, but fun to find out.

About Professor Ueda
Hiroki Ueda graduated from the Faculty of Medicine, the University of Tokyo in 2000, and obtained his Ph.D. in 2004 from the same university. He was appointed as a team leader in RIKEN in 2003. He became a full professor in the Graduate School of Medicine, University of Tokyo in 2013. He is also currently appointed as a team leader in RIKEN Centre for Biosystems Dynamics Research (BDR), an affiliate professor in Graduate School of Information Science and Technology in the University of Tokyo and Osaka and Tokushima University.
In 2016, he found the first sleep-promoting kinases, CaMKIIalpha and CaMKIIbeta and proposed phosphorylation hypothesis of sleep that phosphorylation-dependent regulation of Ca2+-dependent hyperpolarization pathway underlies the regulation of sleep homeostasis in mammals. In 2018, he also found the first essential genes of REM sleep, muscarinic receptors M1 and M3. To accelerate these studies, he also invented whole-brain and whole-body clearing and imaging methods called CUBIC as well as the next-generation mammalian genetics such as Triple-CRISPR and ES-mice methods for one-step production and analysis of KO and KI mice without crossing.

